Implementing FAIR Digital Objects in the EOSC-Life Workflow Collaboratory
Cite as
Carole Goble, Stian Soiland-Reyes, Finn Bacall, Stuart Owen, Alan Williams, Ignacio Eguinoa, Bert Droesbeke, Simone Leo, Luca Pireddu, Laura Rodriguez-Navas, José Mª Fernández, Salvador Capella-Gutierrez, Hervé Ménager, Björn Grüning, Beatriz Serrano-Solano, Philip Ewels, Frederik Coppens (2021):
Implementing FAIR Digital Objects in the EOSC-Life Workflow Collaboratory.
Zenodo (white paper)
https://doi.org/10.5281/zenodo.4605654
Copyright and license
© 2021 Carole Goble et al. Distributed under the terms of Creative Commons Attribution 4.0 international.
Changes by Stian Soiland-Reyes:
- Reformatted as Markdown
- Modified citations to s11 house rules
- URL citations changed to hyperlinks
- Additional hyperlinks for workflow systems
- Reinserted Figure 1
- Remove section “final paper”
- Shorter paragraphs
Implementing FAIR Digital Objects in the EOSC-Life Workflow Collaboratory
Carole Goble¹, Stian Soiland-Reyes¹², Finn Bacall¹, Stuart Owen¹, Alan Williams¹, Ignacio Eguinoa³⁴, Bert Droesbeke³⁴, Simone Leo⁶, Luca Pireddu⁶, Laura Rodriguez-Navas⁷, José Mª Fernández⁷, Salvador Capella-Gutierrez⁷, Hervé Ménager⁸, Björn Grüning⁹, Beatriz Serrano-Solano⁹, Philip Ewels⁵, Frederik Coppens³⁴
¹ Department of Computer Science, The University of Manchester, Manchester, UK
² Informatics Institute, University of Amsterdam, The Netherlands
³ Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
⁴ VIB Center for Plant Systems Biology, Ghent, Belgium
⁵ Science for Life Laboratory (SciLifeLab), Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden
⁶ Center for Advanced Studies, Research and Development in Sardinia (CRS4), Pula, Italy
⁷ Life Sciences Department. Barcelona Supercomputing Center (BSC), Barcelona, Spain
⁸ Pasteur Institute, Paris, France
⁹ Bioinformatics Group, University of Freiburg, Germany
The practice of performing computational processes using workflows has taken hold in the biosciences as the discipline becomes increasingly computational [Reiter 2021]. The COVID-19 pandemic has spotlighted the importance of systematic and shared analysis of SARS-CoV-2 and its data processing pipelines [Hufsky 2020]. This is coupled with a drive in the community towards adopting FAIR practices (Findable, Accessible, Interoperable, and Reusable) not just for data, but also for workflows [Goble 2020], and to improve the reproducibility of processes, both manual and computational.
EOSC-Life brings together 13 of the Life Science ‘ESFRI’ research infrastructures to create an open, digital and collaborative space for biological and medical research. The project is developing a cloud-based workflow collaboratory to drive implementation of FAIR workflows across disciplines and RI boundaries, and foster tool-focused collaborations and reuse between communities via the sharing of data analysis workflows. The collaboratory aims to provide a framework for researchers and workflow specialists to use and reuse workflows. As such it is an example of the Canonical Workflow Frameworks for Research (CWFR) [Hardisty 2020] vision in practice.
EOSC-Life is made up of established research infrastructures ranging from biobanking and clinical trial management, through to coordinating biomedical imaging and plant phenotyping to multi-omic and systems-based data analysis. The heterogeneity of the disciplines is reflected in the diversity of their data analysis needs and practices and the variety of workflow management systems they use. Many have specialist platforms developed over years. Workflow management systems in common use include Galaxy [Afgan 2018], Snakemake [Köster 2012], and Nextflow [Di Tommaso 2017], and more specialist, domain-specific systems such as SCIPION [Gómez-Blanco 2018].
To serve the needs of this established and diverse community, EOSC-Life has developed WorkflowHub as an inclusive workflow registry, agnostic to any Workflow Management System (WfMS). WorkflowHub aims to incorporate their workflows in partnership with the WfMS, to embed the registration of workflows in the community processes, e.g. based on pre-existing workflow repositories.
The registry adopts common practices, e.g. use of GitHub repositories, and supports integration with the ecosystem of tool packages, assisted by registries (bio.tools [Ison 2019], BioContainers [da Veiga Leprevost 2017]), and services for testing and benchmarking workflows (OpenEBench, LifeMonitor) (Figure 1).
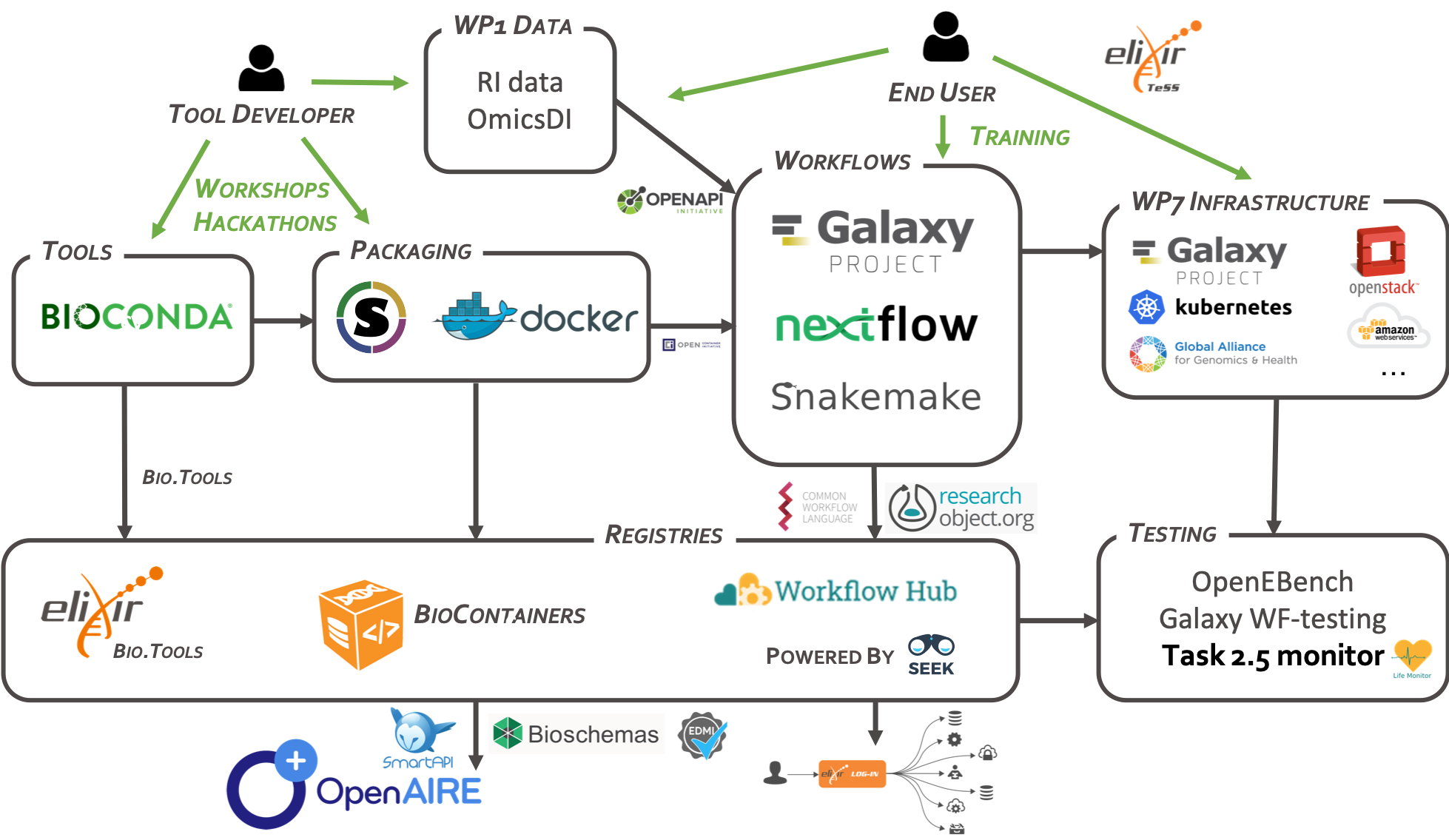
EOSC-Life Collaboratory
Overview of services in the EOSC-Life Collaboratory. Distributed from https://github.com/eosc-life/tools-collaboratory-roadmap under the Apache License, version 2.0.
As an umbrella registry, the Hub makes workflows Findable and Accessible by indexing workflows across workflow management systems and their native repositories, while providing rich standardized metadata. Interoperability and Reusability is supported by standardized descriptions of workflows and packaging of workflow components, developed in close collaboration with the communities.
The WorkflowHub creates a place for registering and discovering libraries of workflows developed by collaborating teams, with suitable features for versioning, credit, analytics, and import/export needed to support the reuse of workflows, the development of sub-workflows as canonical steps and ultimately the identification of common patterns in the workflows.
At the heart of the collaboratory is a Digital Object framework for documenting and exchanging workflows annotated with machine processable metadata produced and consumed by the participating platforms. The Digital Object framework is founded on several needs:
-
Describing a workflow and its steps in a canonical, normalised and WfMS independent way: we use the Common Workflow Language (CWL) [Amstutz 2016], more specifically the Abstract CWL [BioExcel 2020] (non-executable) description variant to accompany the native workflow definitions. This presents the structure, composed tools and external interface in an interoperable way across workflow languages. WfMS can generate abstract CWL, already demonstrated for Galaxy, next to the ‘native’ Galaxy workflow description.
This language duality is an important retention aspect of reproducibility, as the structure and metadata of the workflow can be accessed independent of its native format as CWL, even if that may no longer be executable, capturing the canonical workflow in a FAIR format. The co-presence of the native format enables direct reuse in the specific WfMS, benefitting from all its features.
-
Metadata about a workflow and its tools using a minimal information model: we use the Bioschemas profiles Computational Tool, Computational Workflow and Formal Parameter which are discipline independent, opinionated conventions for using schema.org annotations. Bioschemas enables us to capture and publish workflow registrations and their metadata as FAIR Digital Objects. The EDAM Ontology [Ison 2013] is further used to add bioinformatics-specific metadata, such as strong typing of inputs and outputs, within both Abstract CWL and Bioschemas annotations.
-
Organising and packaging the definitions and components of a workflow with their associated objects such as test data: we use a Workflow profile specialisation of RO-Crate [Ó Carragáin 2019], a community developed standardised approach for research output packaging with rich metadata.
RO-Crate provides us the ability to package executable workflows, their components such as example and test data, abstract CWL, diagrams and their documentation. This makes workflows more readily re-usable. RO-Crate is the base unit of upload and download at the WorkflowHub. As CWFR Digital Objects of workflows, RO-Crates are activation-ready and circulated between the different services for execution and testing.
-
Identifiers for all the components: like FAIR Digital Objects [De Smedt 2020], RO-Crates can be metadata-rich bags of identifiers and can themselves be assigned permanent identifiers. This enables the full description of a computational analysis, from input data, over tools and workflows, to final results.
Using these components we have built an environment that supports the Workflow Life Cycle, from abstract description, through to a specific rendering in a WfMS to its execution and the documentation of its run provenance, results and continued testing.
Funding acknowledgement
This work has received funding from the European Commission’s Horizon 2020 research and innovation programme under grant agreement numbers 824087 (EOSC-Life) and 823830 (BioExcel-2) and is supported by Research Foundation - Flanders (FWO) for ELIXIR Belgium (I002819N).
References
[Afgan 2018] Enis Afgan, Dannon Baker, Bérénice Batut, Marius van den Beek, Dave Bouvier, Martin Čech, John Chilton, Dave Clements, Nate Coraor, Björn Grüning, Aysam Guerler, Jennifer Hillman-Jackson, Vahid Jalili, Helena Rasche, Nicola Soranzo, Jeremy Goecks, James Taylor, Anton Nekrutenko, and Daniel Blankenberg (2018):
The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update.
Nucleic Acids Research 46(W1)
https://doi.org/10.1093/nar/gky379
[Amstutz 2016] Peter Amstutz, Michael R. Crusoe, Nebojša Tijanić (editors), Brad Chapman, John Chilton, Michael Heuer, Andrey Kartashov, Dan Leehr, Hervé Ménager, Maya Nedeljkovich, Matt Scales, Stian Soiland-Reyes, Luka Stojanovic (2016):
Common Workflow Language, v1.0.
Specification, Common Workflow Language working group.
https://doi.org/10.6084/m9.figshare.3115156.v2
[BioExcel 2020] BioExcel (2020):
Creating workflows with Common Workflow Language.
BioExcel Best Practice Guides
https://docs.bioexcel.eu/cwl-best-practice-guide/devpractice/partial.html
[Ó Carragáin 2019] Eoghan Ó Carragáin, Carole Goble, Peter Sefton, Stian Soiland-Reyes (2019):
A lightweight approach to research object data packaging.
Bioinformatics Open Source Conference (BOSC2019)
https://doi.org/10.5281/zenodo.3250687
[Goble 2020] Carole Goble, Sarah Cohen-Boulakia, Stian Soiland-Reyes, Daniel Garijo, Yolanda Gil, Michael R. Crusoe, Kristian Peters, and Daniel Schober (2020):
FAIR Computational Workflows.
Data Intelligence 2(1-2)
https://doi.org/10.1162/dint_a_00033
[Gómez-Blanco 2018] J. Gómez-Blanco, J.M. de la Rosa-Trevín, R. Marabini, L. del Cano, A. Jiménez, M. Martínez, R. Melero, T. Majtner, D. Maluenda, J. Mota, Y. Rancel, E Ramírez-Aportela, J.L. Vilas, M. Carroni, S. Fleischmann, E. Lindahl, A.W. Ashton, M. Basham, D.K. Clare, K. Savage, C.A. Siebert, G.G. Sharov, C.O.S. Sorzano, P. Conesa, J.M. Carazo (2018):
Using Scipion for stream image processing at Cryo-EM facilities.
Journal of Structural Biology 204(3)
https://doi.org/10.1016/j.jsb.2018.10.001
[Hardisty 2020] Alex Hardisty, Peter Wittenburg (eds.) (2020):
Canonical Workflow Framework for Research CWFR- Position Paper V2.
https://osf.io/9e3vc
[Hufsky 2020] Franziska Hufsky, Kevin Lamkiewicz, Alexandre Almeida, Abdel Aouacheria, Cecilia Arighi, Alex Bateman, Jan Baumbach, Niko Beerenwinkel, Christian Brandt, Marco Cacciabue, Sara Chuguransky, Oliver Drechsel, Robert D Finn, Adrian Fritz, Stephan Fuchs, Georges Hattab, Anne-Christin Hauschild, Dominik Heider, Marie Hoffmann, Martin Hölzer, Stefan Hoops, Lars Kaderali, Ioanna Kalvari, Max von Kleist, Renó Kmiecinski, Denise Kühnert, Gorka Lasso, Pieter Libin, Markus List, Hannah F Löchel, Maria J Martin, Roman Martin, Julian Matschinske, Alice C McHardy, Pedro Mendes, Jaina Mistry, Vincent Navratil, Eric P Nawrocki, Áine Niamh O’Toole, Nancy Ontiveros-Palacios, Anton I Petrov, Guillermo Rangel-Pineros, Nicole Redaschi, Susanne Reimering, Knut Reinert, Alejandro Reyes, Lorna Richardson, David L Robertson, Sepideh Sadegh, Joshua B Singer, Kristof Theys, Chris Upton, Marius Welzel, Lowri Williams, Manja Marz (2020):
Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research.
Briefings in Bioinformatics 22(2):bbaa232
https://doi.org/10.1093/bib/bbaa232
[Ison 2013] Jon Ison, Matúš Kalaš, Inge Jonassen, Dan Bolser, Mahmut Uludag, Hamish McWilliam, James Malone, Rodrigo Lopez, Steve Pettifer, Peter Rice (2013):
EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats.
Bioinformatics 29(10)
https://doi.org/10.1093/bioinformatics/btt113
[Ison 2019] Jon Ison, Hans Ienasescu, Piotr Chmura, Emil Rydza, Hervé Ménager, Matúš Kalaš, Veit Schwämmle, Björn Grüning, Niall Beard, Rodrigo Lopez, Severine Duvaud, Heinz Stockinger, Bengt Persson, Radka Svobodová Vařeková, Tomáš Raček, Jiří Vondrášek, Hedi Peterson, Ahto Salumets, Inge Jonassen, Rob Hooft, Tommi Nyrönen, Alfonso Valencia, Salvador Capella, Josep Gelpí, Federico Zambelli, Babis Savakis, Brane Leskošek, Kristoffer Rapacki, Christophe Blanchet, Rafael Jimenez, Arlindo Oliveira, Gert Vriend, Olivier Collin, Jacques van Helden, Peter Løngreen & Søren Brunak (2019):
The bio.tools registry of software tools and data resources for the life sciences.
Genome Biology 20:164
https://doi.org/10.1186/s13059-019-1772-6
[Köster 2012] Johannes Köster, Sven Rahmann (2012):
Snakemake—a scalable bioinformatics workflow engine.
Bioinformatics 28(19)
https://doi.org/10.1093/bioinformatics/bts480
[Reiter 2021] Taylor Reiter, Phillip T Brooks, Luiz Irber, Shannon E K Joslin, Charles M Reid, Camille Scott, C Titus Brown, N Tessa Pierce-Ward (2021):
Streamlining data-intensive biology with workflow systems.
GigaScience 10(1):giaa140
https://doi.org/10.1093/gigascience/giaa140
[De Smedt 2020] Koenraad De Smedt, Dimitris Koureas, Peter Wittenburg (2020):
FAIR Digital Objects for Science: From Data Pieces to Actionable Knowledge Units.
Publications 8(2):21
https://doi.org/10.3390/publications8020021
[Di Tommaso 2017] Paolo Di Tommaso, Maria Chatzou, Evan W Floden, Pablo Prieto Barja, Emilio Palumbo, Cedric Notredame (2017):
Nextflow enables reproducible computational workflows.
Nature Biotechnology 35(4)
https://doi.org/10.1038/nbt.3820
[da Veiga Leprevost 2017] Felipe da Veiga Leprevost, Björn A Grüning, Saulo Alves Aflitos, Hannes L Röst, Julian Uszkoreit, Harald Barsnes, Marc Vaudel, Pablo Moreno, Laurent Gatto, Jonas Weber, Mingze Bai, Rafael C Jimenez, Timo Sachsenberg, Julianus Pfeuffer, Roberto Vera Alvarez, Johannes Griss, Alexey I Nesvizhskii, Yasset Perez-Riverol (2017):
BioContainers: an open-source and community-driven framework for software standardization.
Bioinformatics 33(16)
https://doi.org/10.1093/bioinformatics/btx192